Single Cell ATAC-Seq Service
Single-cell ATAC Sequencing (short for “Assay for Transposase Accessible Chromatin”) is a technique used to determine genome-wide chromatin accessibility at the single-cell level. With single-cell ATAC-Seq, chromatin profiling of thousands of nuclei can be performed in parallel, resulting in fast, accurate epigenomic profiling.
While bulk ATAC-seq has proven a powerful method for determining the broad characteristics of a population, tissues and other biological samples tend to contain multiple cell types in varying states of cellular processes. To capture this variation, it is necessary to sequence individual cells.
Using the industry-leading Chromium Single Cell ATAC Solution from 10x Genomics, SingulOmics’ PhD-level scientists and bioinformaticians can analyze hundreds to thousands of nuclei per run at single-cell resolution. Our technical team includes leading scientists who have made important contributions to the single-cell field. Our comprehensive sequencing and analysis solutions have multiple applications, including epigenetic regulatory networks, tumor heterogeneity, and biomarker discovery. In combination with Single-Cell RNA-Seq, we also provide 10x Single Cell Multiome ATAC + Gene Expression service for simultaneous profiling of gene expression and open chromatin from the same cell, enabling researchers to link input regulatory signals with their output gene expression at the single-cell level for thousands of cells.
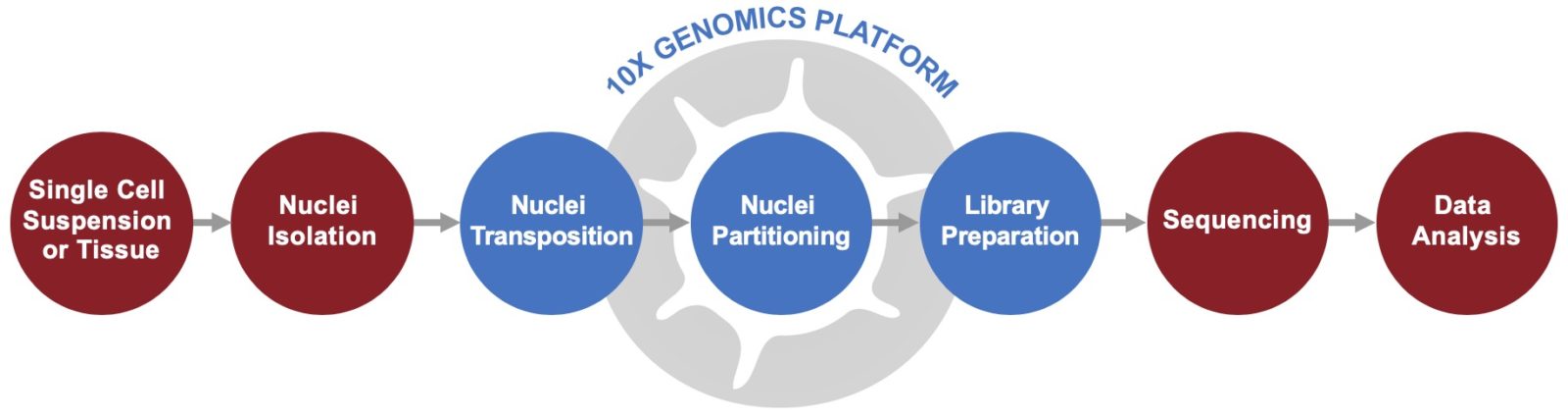
Service Workflow
Sample Requirements
Sample requirements for frozen cells
(1) We recommend freezing 5x10^5 to 1x10^6 cells in 1 ml of freezing media per cryotube.
(2) If the number of cells is not a limiting factor, freeze 2x10^6 cells in 1 ml of freezing media per cryotube and prepare two (2) cryotubes per sample. If you have a limited number of cells, you can still submit 1x10^5 cells in 0.5 ml of freezing media per cryotube.
(3) Use standard freezing media (e.g. cell growth media + 20% FBS + 10% DMSO; or 90% FBS + 10% DMSO) and place the samples at -80°C while slowly decreasing the temperature using any device/method to maintain an approximate -1°C/minute freeze rate.
Sample requirements for frozen tissue (for nuclei isolation)
Fresh tissue, immediate after collection, must be "snap" or "flash" frozen on dry ice or in liquid nitrogen to preserve RNA integrity. Failure to do so will affect the final quality of the isolated nuclei.
(1) If possible, smaller size tissue (approx. 100 mg) is preferred as it can be frozen faster than a larger one.
(2) One method we recommend is using Dry Ice Ethanol Bath. Always wear gloves when handling dry-ice and ethanol. Break the dry ice into smaller pieces and place them into a rubber or stainless steel container (plastic can degrade over time). Pour the ethanol solution into the container; add only enough to completely cover the dry ice. Once the boiling ethanol solution has slowed down, place your samples in.
Before submitting your samples, contact us to discuss the best approach for your particular cell type.
Next Steps...
Contact us to learn more or get a price quote.
