End-to-end Bulk RNA-Seq Service with Cell Type Deconvolution
Bulk RNA sequencing (RNA-Seq) has been the method of choice for profiling transcriptomic variations under different conditions such as disease states. Bulk RNA-Seq measures the average gene expression levels by summing over the population of cells in complex tissues with heterogeneous cell types.
Certain cell types are more vulnerable to diseases, and the expression level of each gene varies among different cell types. Gene expressions are affected by clinical/biological conditions and cell type proportion differences among samples. However, gene expression analyses of bulk tissues often ignore cell type composition as an important confounding factor, resulting in loss of signal from rare cell types and bias towards dominant cell types.
Using cell-type-specific gene expression references from single-cell studies, we can now characterize the variation of cell-type composition/proportions for disease-relevant tissues by in silico deconvolution of bulk RNA-Seq data. We can also identify genes differentially expressed due to clinical/biological conditions without bias from cell type proportion differences.
Case Study: Cell Type Deconvolution of PBMC Samples
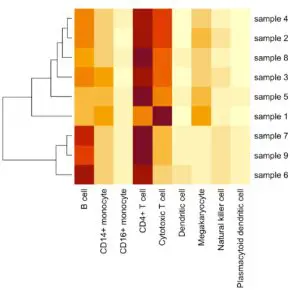
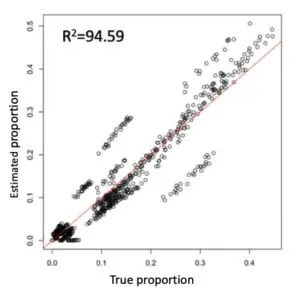
We constructed a single-cell gene expression reference by combining four 10x Chromium datasets from two PBMC samples (Ding et al., Nature Biotechnology 2020). The single-cell reference contains 13,028 cells, covering nine cell types. We then tested our deconvolution pipeline using pseudo-bulk RNA-Seq samples whose true underlying cell-type identities are known. The predicted cell-type proportions and the actual proportions are highly consistent.
We can deconvolute any tissue type using the corresponding single cell reference(s).
Service Workflow
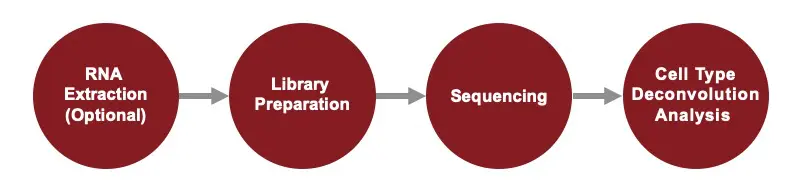
Deconvolution analysis includes (1) characterization of cell-type composition, and (2) differential gene expression analysis without bias from cell-type proportion differences among samples.
RNA sequencing is done at 2 x 150 bp with a data quality guarantee (Q30 ≥ 80%).
Minimum sample number: 24 samples
Next Steps...
Contact us to learn more or get a price quote.
