
10x Visium Spatial Transcriptomics Service
Spatially resolved transcriptomic technologies are unlocking novel insights into tissue biology. With the ability to measure gene expression with spatial context, you can map the organization of cell types and states in tissue and uncover their interactions and relationships. This information can yield valuable information for developmental biology, disease pathology, and clinical and translational research.
At SingulOmics, we offer spatial transcriptomic analysis using the Visium Spatial Gene Expression workflow from 10x Genomics. Visium Spatial Gene Expression maps the whole transcriptome in entire tissue sections with morphological context and high cellular resolution. We also enable spatial multiomics analysis using the Visium Spatial Gene Expression with Immunofluorescence (IF) solution. Combine the power of whole transcriptome spatial analysis with IF protein co-detection from the same sample for a more detailed picture of the tissue microenvironment.
Our expert team is highly experienced with single-cell methods and histology and offers a comprehensive service workflow covering every step from tissue sectioning to data analysis.
Spatial Gene Expression for Fresh Frozen Tissue
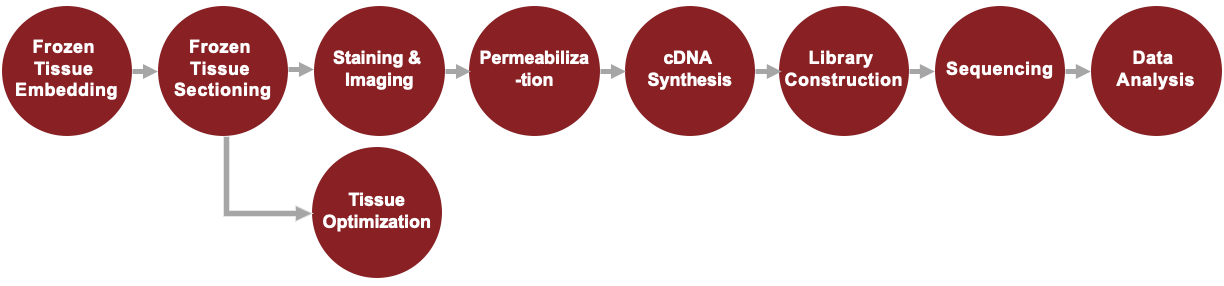
Visium Spatial Gene Expression uses slides with capture areas containing thousands of barcoded spots, each containing millions of capture oligonucleotides. Tissues are sectioned and placed on the capture areas, then stained with H&E or immunofluorescence. Permeabilizing the tissue releases mRNA which then binds to spatially barcoded capture spots. Tissue optimization is performed to determine the best tissue permeabilization condition prior to the permeabilization step. cDNA is produced from the captured mRNA and used to generate a sequencing library. After sequencing, comprehensive data analysis is performed to overlay gene expression onto the histological image and identify marker genes and cell types in the context of the heterogenous tissue microenvironment.
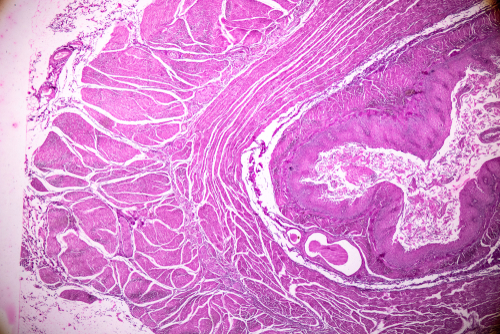
Spatial Gene Expression for FFPE Tissue
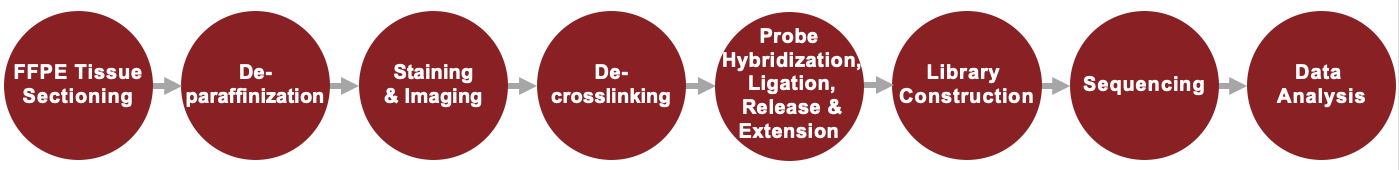
Formalin-fixed paraffin-embedded (FFPE) tissue is a common tissue preservation method but represents unique challenges for transcriptomic detection as FFPE processing damages RNA. The separate Visium for FFPE assay overcomes these hurdles using a split-probe approach and RNA-templated ligation. FFPE tissues are sectioned and placed on the capture areas, deparaffinized, stained with H&E or immunofluorescence, and de-crosslinked. This is followed by probe hybridization, probe ligation, and probe release and extension. The spatially barcoded, ligated probe products are released from the slide and used for library construction. After sequencing, comprehensive data analysis allows users to map the whole transcriptome in FFPE tissue.

We use the Visium CytAssist instrument to automate the transfer of analytes from standard histology glass slides to Visium slides, expanding the range of samples we accept to FFPE blocks, pre-sectioned tissues on glass slides, and archived tissue samples. By pre-screening for biologically significant tissue sections, we ensure high-quality results from the Visium assay.
Visium HD Spatial Gene Expression
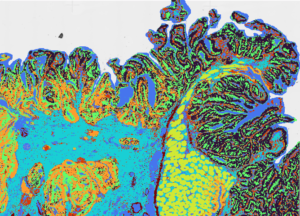 Singulomics now offers the Visium HD Spatial Gene Expression service, enabling comprehensive single-cell scale analysis of FFPE and frozen tissue sections. Utilizing CytAssist technology, Visium HD maps whole transcriptome mRNA expression with unmatched precision, using millions of 2 x 2 µm barcoded squares for continuous tissue coverage. This innovative tool allows for detailed tissue characterization, integrating with H&E or IF imaging to enhance histological insights. With our expertise in histology, spatial transcriptomics, and bioinformatics, Singulomics efficiently processes large and complex datasets, delivering publication-ready results that bring your research to the forefront of spatial transcriptomics.
Singulomics now offers the Visium HD Spatial Gene Expression service, enabling comprehensive single-cell scale analysis of FFPE and frozen tissue sections. Utilizing CytAssist technology, Visium HD maps whole transcriptome mRNA expression with unmatched precision, using millions of 2 x 2 µm barcoded squares for continuous tissue coverage. This innovative tool allows for detailed tissue characterization, integrating with H&E or IF imaging to enhance histological insights. With our expertise in histology, spatial transcriptomics, and bioinformatics, Singulomics efficiently processes large and complex datasets, delivering publication-ready results that bring your research to the forefront of spatial transcriptomics.
Service Workflow
Visium Spatial Gene Expression for Fresh Frozen Tissue Service Workflow
Visium Spatial Gene Expression for FFPE Service Workflow
Sample Requirements
We accept the following sample types for the Fresh Frozen workflow:
· Flash frozen tissue embedded in OCT
· Flash frozen tissue without OCT
We accept the following sample types for the FFPE workflow:
· FFPE blocks (human or mouse only)
· Pre-sectioned tissue on glass sides (human or mouse only)
Next Steps...
Contact us to learn more or get a price quote.
