High-Throughput or Deep Single Cell RNA-Seq Service
Single cell RNA-Seq opens a whole new field in biology by allowing the study of cell-to-cell transcriptome heterogeneity. It enables the discovery of cellular differences typically masked by standard, bulk RNA sequencing. Built on a foundation of unmatched expertise in single-cell genomics, transcriptomics and epigenomics developed since mid-2000s, SingulOmics is highly experienced in single cell RNA sequencing and data analysis.
High-Throughput 10x Genomics Chromium Single Cell RNA-Seq
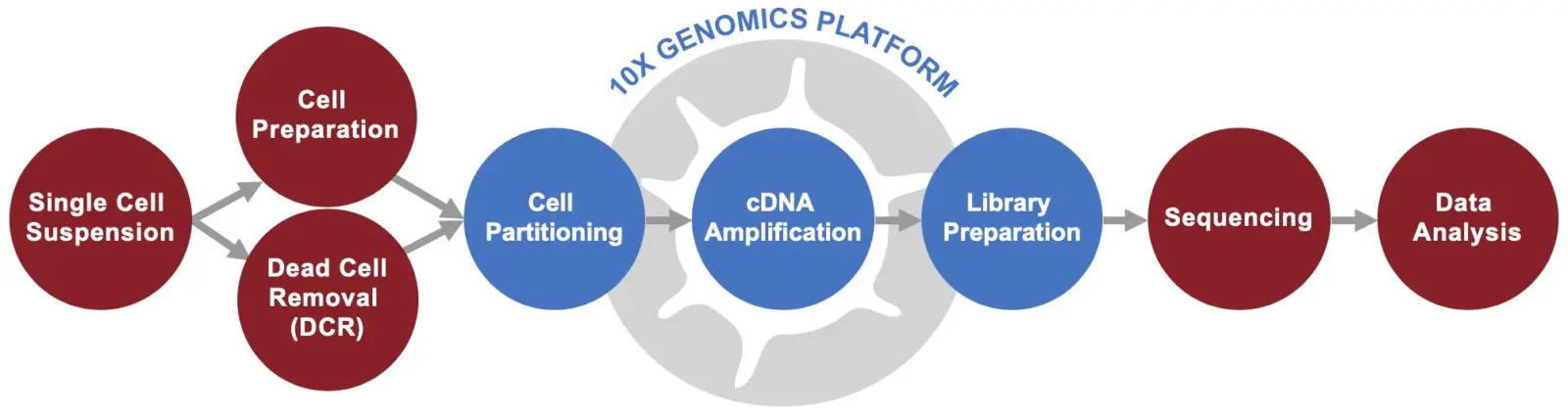
We provide high-throughput single cell gene expression profiling service using 10x Genomics Chromium Platform to examine thousands of cells per sample through cell-by-cell 3’ end counting of mRNA transcripts. Our end-to-end service solution enables the identification of rare cell types, analysis of cellular heterogeneity, and understanding of individual cell states and cellular responses to environmental signals and conditions.
To ensure high-quality results, SingulOmics has optimized our experimental workflow to include Dead Cell Removal. The addition of this important step increases cell viability, thus improving cell recovery and ensuring high sequencing accuracy. Our scientists are highly experienced in performing dead cell removal and can determine if your samples should undergo this process to provide the best possible results.
10x Genomics Chromium Single Nucleus RNA-Seq
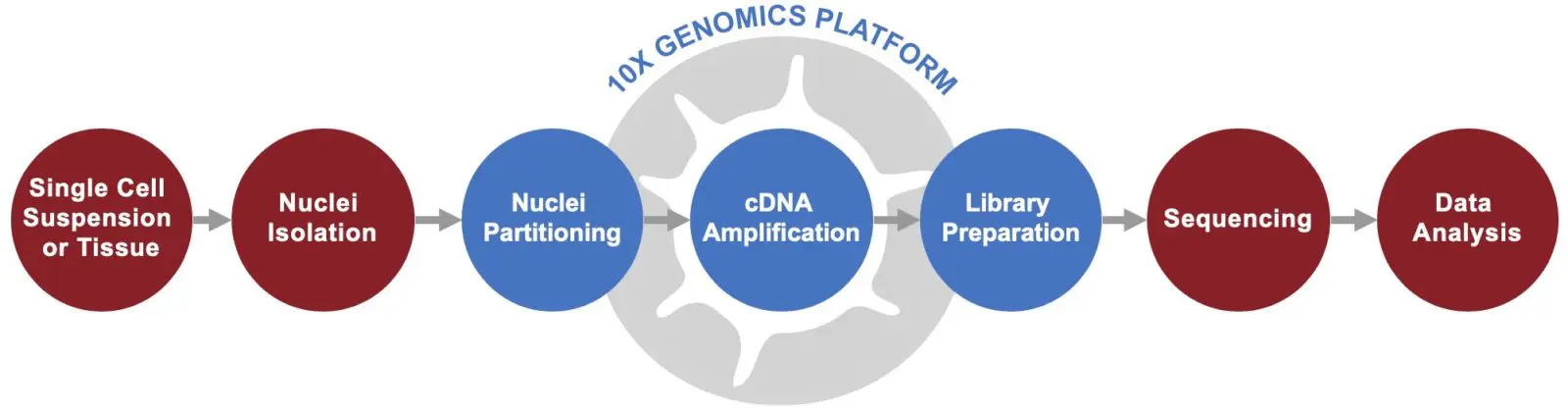
Depending on the project type, single nuclei may be a better fit as sample input. This is common when studying tissue samples from which it is difficult to extract intact cells, such as brain tissue or flash frozen tissue. Singulomics has highly-skilled scientists who have experience conducting nuclei isolation from flash frozen tissues and can generate single-nuclei suspensions as input for 10x Genomics Chromium single-cell gene expression profiling.
In addition, we are experienced in handling small amounts of frozen tissue, such as a tissue biopsy. We have successfully conducted 10x single nucleus RNA-Seq for a wide variety of tissue types from multiple species, including mammals, amphibians, reptiles, and insects.
10x Genomics Chromium Single Cell Immune Profiling
Comprehensively profile the immune system at single cell resolution for clonotyping studies, large-scale antibody discovery, and more. Using Chromium, Single Cell Immune Profiling, we generate two or three libraries from a single input sample for:
· 5’ gene expression analysis
· T cell receptor repertoire profiling
· B cell immunoglobulin repertoire profiling
These libraries are sequenced and analyzed integratively in downstream informatics analysis.
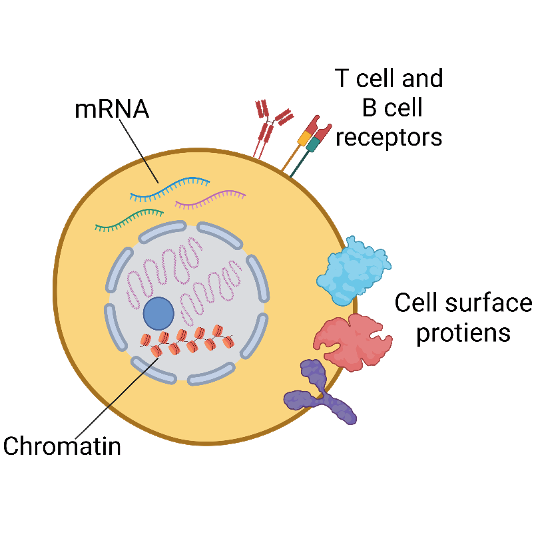
10x Single Cell RNA-Seq with Cell Surface Protein (CITE-Seq)
The CITE-Seq (Cellular Indexing of Transcriptomes and Epitopes by Sequencing) method enables integrated protein and transcriptome measurement with single-cell resolution. We can label cell surface proteins with DNA-barcoded antibodies to enable combined gene expression profiling and cell surface protein detection from the same cell for Single Cell RNA-Seq and Immune Profiling services.
10x Single Cell Multiome ATAC + Gene Expression
Discover how genes are expressed and regulated using our 10x Single Cell Multiome ATAC + Gene Expression service. With simultaneous profiling of gene expression and open chromatin from the same cell, you can link input regulatory signals with their output gene expression at the single-cell level for thousands of cells. We leverage years of experience in nuclei isolation from cryopreserved cells and flash frozen tissues to provide accurate, high-quality data.
10x Single Cell Flex Gene Expression for Whole Blood
The 10x Chromium Single Cell Flex Gene Expression workflow—also known as Fixed RNA Profiling—enables high-quality single-cell RNA-seq from fixed whole blood samples. This method preserves cellular states at the time of blood collection and allows storage at −80ºC for up to one month, supporting batch shipping and flexible processing. At Singulomics, we specialize in isolating PBMCs or leukocytes from fixed blood, with protocols optimized to preserve fragile neutrophils. Our end-to-end workflow delivers consistent and reliable gene expression data for comprehensive whole blood transcriptomic analysis.

10x Single Cell Flex Gene Expression for FFPE Tissue Samples
Unlock the transcriptomic potential of archived tissues with our complete service using 10x Chromium Single Cell Flex Gene Expression (also known as Fixed RNA Profiling). From tissue block to transcriptomic insight, we provide end-to-end support—including sectioning, deparaffinization, decrosslinking, section dissociation to single cells and nuclei, library preparation, sequencing, and data analysis. Our optimized workflow delivers consistent and reliable gene expression data from preserved tissues previously considered incompatible with single-cell analysis, opening new possibilities for translational and clinical research.
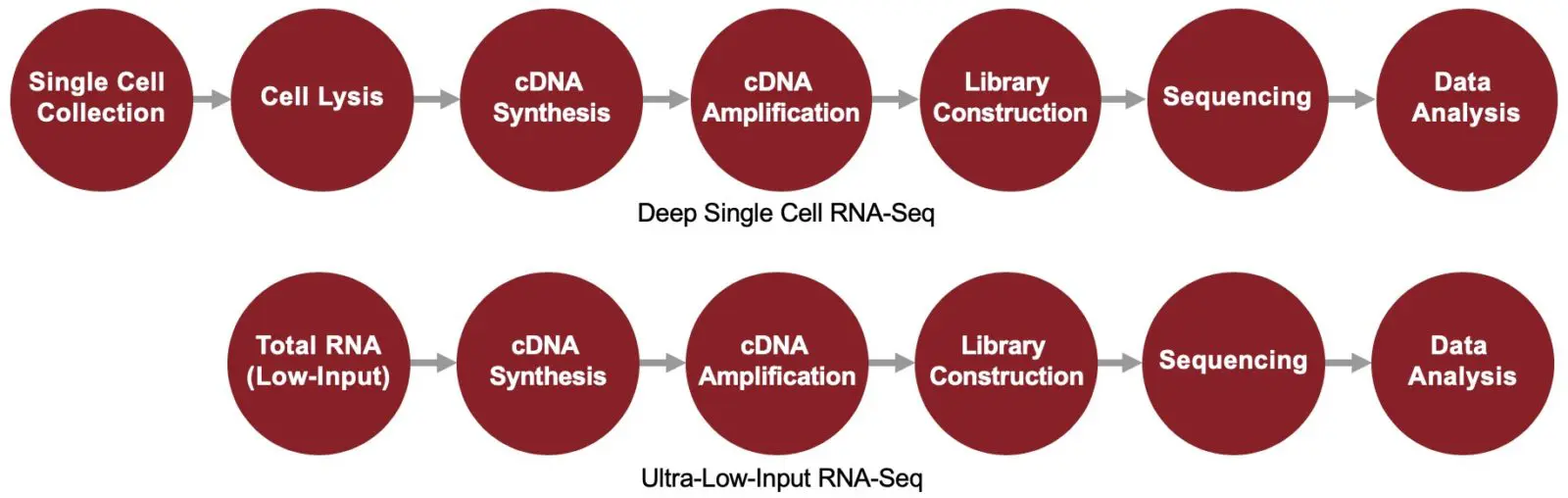
Deep Single Cell / Ultra-Low-Input RNA-Seq
To enable the discovery of transcriptomes and gene expressions of single cells at a deeper level, SingulOmics offers deep single cell RNA-Seq service. The same technology can also be applied to samples with limited number of cells (1-1000 cells) or with ultra-low amount of input RNA (ultra-low-input RNA-Seq).
Service Workflow
High-Throughput 10x Genomics Single Cell RNA-Seq
Ultra-Low-Input / Deep Single Cell RNA-Seq
Sample Requirements
High-Throughput 10x Genomics Single Cell RNA-Seq
Sample requirements for frozen cells
(1) We recommend freezing 5x10^5 to 1x10^6 cells in 1 ml of freezing media per cryotube.
(2) If the number of cells is not a limiting factor, freeze 2x10^6 cells in 1 ml of freezing media per cryotube and prepare two (2) cryotubes per sample. If you have a limited number of cells, you can still submit 1x10^5 cells in 0.5 ml of freezing media per cryotube.
(3) Use standard freezing media (e.g. cell growth media + 20% FBS + 10% DMSO; or 90% FBS + 10% DMSO) and place the samples at -80°C while slowly decreasing the temperature using any device/method to maintain an approximate -1°C/minute freeze rate.
Sample requirements for frozen tissue (for nuclei isolation)
Fresh tissue, immediate after collection, must be "snap" or "flash" frozen on dry ice or in liquid nitrogen to preserve RNA integrity. Failure to do so will affect the final quality of the isolated nuclei.
(1) If possible, smaller size tissue (approx. 100 mg) is preferred as it can be frozen faster than a larger one.
(2) One method we recommend is using Dry Ice Ethanol Bath. Always wear gloves when handling dry-ice and ethanol. Break the dry ice into smaller pieces and place them into a rubber or stainless steel container (plastic can degrade over time). Pour the ethanol solution into the container; add only enough to completely cover the dry ice. Once the boiling ethanol solution has slowed down, place your samples in.
Before submitting your samples, contact us to discuss the best approach for your particular cell type.
Ultra-Low-Input / Deep Single Cell RNA-Seq
Samples can be submitted as (1) single cells (nuclei), (2) frozen cells in cell suspensions, or (3) total RNA.
(1) Single cells (nuclei): Please click here to access instructions on how to collect single cells.
(2) Frozen cells of single population in cell suspensions: We provide single cell isolation service using CellRaftTM. Please click here for sample requirements.
If you have additional requirements for single cell isolation (e.g. single cell population from blood or tissues), please contact us.
(3) Total RNA, please click here for sample requirements.
Next Steps...
Contact us to learn more or get a price quote.
